My old videos about analytical separation processes should be added to this site. The scripts are still good and easy to modify to simulate different scenario by changing a few parameters.
When I first made them in 1996 and 1999, I saved them in mpeg1 formats. Nowadays, other formats are more popular, especially MP4 that can run on practically 100% of the browsers including iPhone and iPad — and on any other browser — since Flash players can handle that one as well. Just because I like the native Firefox format, I added OGV files as well. The MATLAB scripts were set to dump uncompressed AVI files. The AVI files were converted to MP4 using Mpegstreamclip, and to OGV using fffmpeg2theora. The below code can be copied and modified in MATLAB.
Open tubular chromatography simulation
% Create an empty dark window
figure(1);
set(1,‘Position’,[69 601 620 66]);
clf;
colormap(‘gray’)
fill([0 0 5000 5000 0],[-200 200 200 -200 -200],‘k’)
% Video picture, frame 1
axis([0 5000 -140 140]); axis(‘equal’); axis(‘off’);
text(500,100,‘Made by Martin Andersson, Dept. of Technical Analytical Chemistry’,‘Color’,‘w’)
text(1500,-50,‘Lund University, Sweden 1996’,‘Color’,‘w’)
mov=avifile(‘cromat.avi’,‘compression’,‘none’);
% Parameters for the simulation
flow=20; % Speed of the plug flow
diffusion=10; % Diffusionen
pA=0.2; % partition OR propability for analyte A
pB=1; % partition OR propability for analyte B
N=30; % Number of molecules of each analyte
% Set up the plug of sample, “inject” the sample
Startx=ones(10,1)*[20:20:200];
Starty=[-90:20:90]’*ones(1,10)*diag([1 -1 1 -1 1 -1 1 -1 1 -1]);
Apos=[Startx(1:2:N*2)’ Starty(1:2:N*2)’];
Bpos=[Startx(2:2:N*2)’ Starty(2:2:N*2)’];
% Draw the injected sample
hold off;
fill([0 0 5000 5000 0],[-200 -100 -100 -200 -200],‘y’)
hold on;
fill([0 0 5000 5000 0],[100 200 200 100 100],‘y’)
fill([0 0 5000 5000 0],[-100 100 100 -100 -100],‘w’);
plot(Apos(:,1),Apos(:,2),‘r.’,‘Markersize’,10);
plot(Bpos(:,1),Bpos(:,2),‘b.’,‘Markersize’,10);
axis([0 5000 -140 140]); axis(‘equal’); axis(‘off’);
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
% t>0. Flow starts.
count=2;
while count count=count+1;
% Move analytes A and B forward that are not attached to the wall
mobileA=find(abs(Apos(:,2)) Apos(mobileA,:)=Apos(mobileA,:)+diffusion*randn(size(Apos(mobileA,:)));
Apos(mobileA,1)=Apos(mobileA,1)+flow;
mobileB=find(abs(Bpos(:,2)) Bpos(mobileB,:)=Bpos(mobileB,:)+diffusion*randn(size(Bpos(mobileB,:)));
Bpos(mobileB,1)=Bpos(mobileB,1)+flow;
% If stuck at the walls, released according to probability of A and B
ind=find(Apos(:,2)> 80); Apos(ind,2)=130*ones(size(ind));
ind=ind(rand(size(ind))>pA); Apos(ind,2)=80*ones(size(ind));
ind=find(Apos(:,2) ind=ind(rand(size(ind))>pA); Apos(ind,2)=-90*ones(size(ind));
ind=find(Bpos(:,2)> 80); Bpos(ind,2)=130*ones(size(ind));
ind=ind(rand(size(ind))>pB); Bpos(ind,2)=80*ones(size(ind));
ind=find(Bpos(:,2) ind=ind(rand(size(ind))>pB); Bpos(ind,2)=-90*ones(size(ind))
% Draw walls and decorate
hold off;
fill([0 0 5000 5000 0],[-200 -100 -100 -200 -200],‘y’)
hold on;
fill([0 0 5000 5000 0],[100 200 200 100 100],‘y’)
fill([0 0 5000 5000 0],[-100 100 100 -100 -100],‘w’);
% Plot the sample
plot(Apos(:,1),Apos(:,2),‘r.’,‘Markersize’,10);
plot(Bpos(:,1),Bpos(:,2),‘b.’,‘Markersize’,10);
axis([0 5000 -140 140]); axis(‘equal’); axis(‘off’);
figure(gcf);
drawnow;
frame=getframe(gcf);
mov=addframe(mov,frame);
end;
mov=close(mov);
Field flow fractionation separation simulation
% Parameters
CrossFlow=-1;
Sdiffusion=2 % diffusion of Small analyte;
Ldiffusion=1; % diffusion of Large analyte
LaminarConst=1.5; %LINEAR flow profile, “slope”=1.5
N=50; %Number of molecules of each analyte
Width=100; % Channel width
Length=1000; % Channel length
%Print-size of molecules
markerSizeL = 5*3;
markerSizeS = 2*3;
mov=avifile(‘fff.avi’,‘compression’,‘none’);
% t=0. Inject sample at middle of top.
% Large and Small positions.
Lpos=[ones(N,1)*Width/2 ones(N,1)*Width];
Spos=[ones(N,1)*Width/2 ones(N,1)*Width];
hold off;
plot([Lpos(:,1) ones(N,1)*NaN]’, [Lpos(:,2) ones(N,1)*NaN]’,’.m’,‘MarkerSize’,markerSizeL);
hold on;
plot([Spos(:,1) ones(N,1)*NaN]’, [Spos(:,2) ones(N,1)*NaN]’,’.y’,‘MarkerSize’,markerSizeS);
set(gcf,‘position’,[45 453 620 200]);
axis([-Width Length -5 Width]);
set(gca,‘ytick’,[]);
set(gca,‘xtick’,[]);
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
% t>0. Crossflow starts. Relaxation & focusing.
count=1;
mL=Width;
mS=Width;
maxS=Inf;
while maxS>0.15*Width,
count=count+1
Lpos=Lpos+Ldiffusion*randn(size(Lpos));
Lpos(:,2)=Lpos(:,2)+CrossFlow;
Spos=Spos+Sdiffusion*randn(size(Spos));
Spos(:,2)=Spos(:,2)+CrossFlow;
% Bounce at the “walls” — focusing using tentative walls.
ind=find(Lpos(:) ind=find(Spos(:) ind=find(Lpos(:)>Width+1); Lpos(ind)=Width+1-(Lpos(ind)-(Width+1));
ind=find(Spos(:)>Width+1); Spos(ind)=Width+1-(Spos(ind)-(Width+1));
hold off;
plot([Lpos(:,1) ones(N,1)*NaN]’, [Lpos(:,2) ones(N,1)*NaN]’,’.m’,‘MarkerSize’,markerSizeL);
hold on;
plot([Spos(:,1) ones(N,1)*NaN]’, [Spos(:,2) ones(N,1)*NaN]’,’.y’,‘MarkerSize’,markerSizeS);
maxS=max(Spos(:,2));
plot([-Width Length],[0 0],‘w-’)
set(gca,‘ytick’,[]);
set(gca,‘xtick’,[]);
axis([-Width Length -5 Width]);
text(0.2*Length,0.8*Width, ‘Relaxation/focusing. Crossflow only’);
drawnow;
frame=getframe(gcf);
mov=addframe(mov,frame);
end;
% separation after focusing
while count count=count+1
Lpos=Lpos+Ldiffusion*randn(size(Lpos));
Lpos(:,2)=Lpos(:,2)+CrossFlow;
Lpos(:,1)=Lpos(:,1)+(Lpos(:,2)+2)*LaminarConst;
Spos=Spos+Sdiffusion*randn(size(Spos));
Spos(:,2)=Spos(:,2)+CrossFlow;
Spos(:,1)=Spos(:,1)+(Spos(:,2)+2)*LaminarConst;
% Bounce at the wall.
ind=find(Lpos(:) ind=find(Spos(:) hold off;
plot([Lpos(:,1) ones(N,1)*NaN]’, [Lpos(:,2) ones(N,1)*NaN]’,’.m’,‘MarkerSize’,markerSizeL);
hold on;
plot([Spos(:,1) ones(N,1)*NaN]’, [Spos(:,2) ones(N,1)*NaN]’,’.y’,‘MarkerSize’,markerSizeS);
% Draw distribution curves…
% mL=[mean(Lpos(:,1)) mL(1)];
% mS=[mean(Spos(:,1)) mS(1)];
% sL=std(Lpos(:,1));
% sS=std(Spos(:,1));
% NL=exp(-([-Width:Length]-mL(1)).^2/2/sL/N)/sL;
% NS=exp(-([-Width:Length]-mS(1)).^2/2/sS/N)/sS;
% plot(-Width:Length,NL*1000,‘w-’)
% plot(-Width:Length,NS*2000,‘w:’)
% plot(mL(1)*[1 1],[0 Width],’-w’);
% plot(mS(1)*[1 1],[0 Width],’:w’);
Length],[0 0],‘w-’)
set(gca,‘ytick’,[]);
set(gca,‘xtick’,[]);
axis([-Width Length -5 Width]);
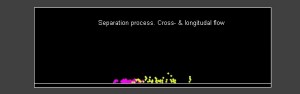
text(0.2*Length,0.8*Width,‘Separation process. Cross- & longitudal flow’);
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end;
mov=close(mov);
disp(‘done’);
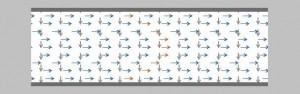
Field flow fractionation, demonstrating the flows
Download mat-file with the arrow images to be loaded before running this script.
step=[5.0 6 6.5 7 7.125];
mov=avifile(‘fl-fff.avi’,‘compression’,‘none’);
% Show the laminar flow
for k=1:120,
shift=round(step*k);
for k=1:length(step);
r{k}=[r1(:,end-shift(k)+1:end,:) r1(:,1:end-shift(k),:)];
end
xx=[r{1};r{2};r{3};r{4};r{5};r{4};r{3};r{2};r{1}];
xx=[zeros(16,960,3)+128; xx;zeros(16,960,3)+128];
if k==1, % make a nice transition
for f=0:0.05:0.95,
imshow(uint8(double(xx(1:2:end,1:2:end,:))*f+0*(1-f)));
drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end
end
imshow(xx(1:2:end,1:2:end,:));
if k drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end
for f=0.05:0.05:1, % make a nice transition
imshow(uint8(double(xx(1:2:end,1:2:end,:))*(1-f)+255*f));
drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end
% Show the cross flow
for k=1:90,
ccc=cc([end‑k+1:end 1:end‑k],:,:);
ccc=[zeros(16,960,3)+128; ccc;zeros(16,960,3)+128];
if k==1,% make a nice transition
for f=0:0.05:0.95,
imshow(uint8(double(ccc(1:2:end,1:2:end,:))*f+255*(1-f)));
drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end
end
imshow(ccc(1:2:end,1:2:end,:));
if k drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end
for f=0.05:0.05:1,% make a nice transition
imshow(uint8(double(ccc(1:2:end,1:2:end,:))*(1-f)+255*f));
drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end
%Show both
for k=1:120,
shift=round(step*k);
for k=1:length(step);
r{k}=[r1(:,end-shift(k)+1:end,:) r1(:,1:end-shift(k),:)];
end
xx=[r{1};r{2};r{3};r{4};r{5};r{4};r{3};r{2};r{1}];
xx=[zeros(16,960,3)+128; xx;zeros(16,960,3)+128];
ccc=cc([end‑k+1:end 1:end‑k],:,:);
ccc=[zeros(16,960,3)+128; ccc;zeros(16,960,3)+128];
xc=min(xx,ccc);
if k==1,% make a nice transition
for f=0:0.05:0.95,
imshow(uint8(double(xc(1:2:end,1:2:end,:))*f+255*(1-f)));
drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end,
end
imshow(xc(1:2:end,1:2:end,:));
if k drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end
for f=0:0.05:0.95,% make a nice transition
imshow(uint8(double(xc(1:2:end,1:2:end,:))*(1-f)+0*f));
drawnow;
figure(gcf);
frame=getframe(gcf);
mov=addframe(mov,frame);
end
mov=close(mov);